Hailing Shi Lab at Emory University School of Medicine aims to combine nucleic acid chemistry, spatial and single-cell multi-omics, and machine-learning-assisted computation to enable characterization and modulation tools for single-cell and context-resolved RNA biology. The overarching goal is to understand and ultimately engineer cell plasticity to benefit health and treat diseases in the brain and other important organs.
Spatial RNA Biology
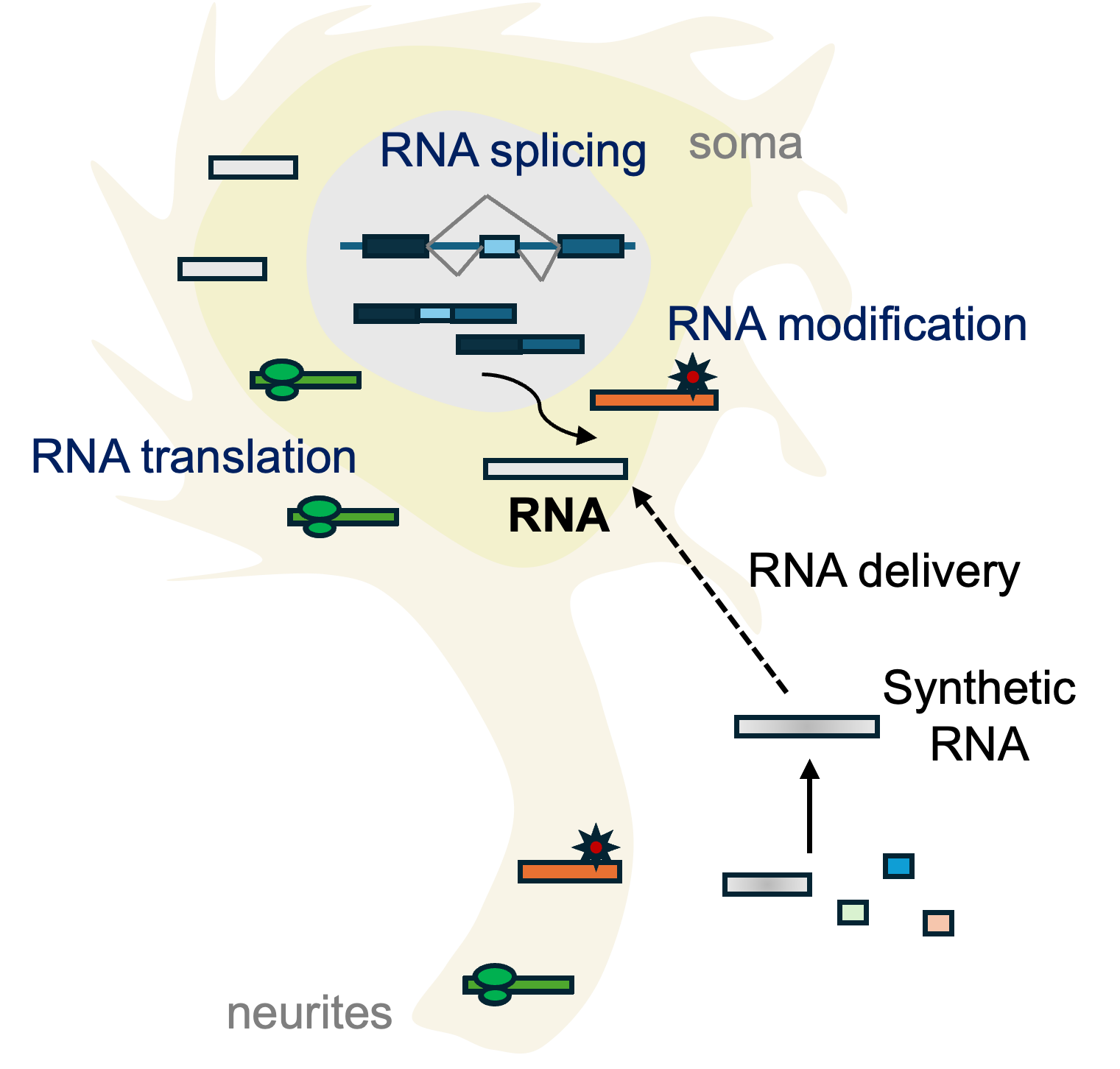
Gene expression is extensively regulated at the RNA level in the nervous system during development and function, including abundant RNA modifications, alternation splicing, circular RNA formation, etc. In addition, RNA binding protein dysregulation has also been observed in various neurodegenerative and neurological disorders.
However, the brain is a spatially organized tissue comprising diverse local cellular environments and extended cell morphologies. It remains challenging to understand RNA biology in cell-type and subcellular-compartment-resolved matters.
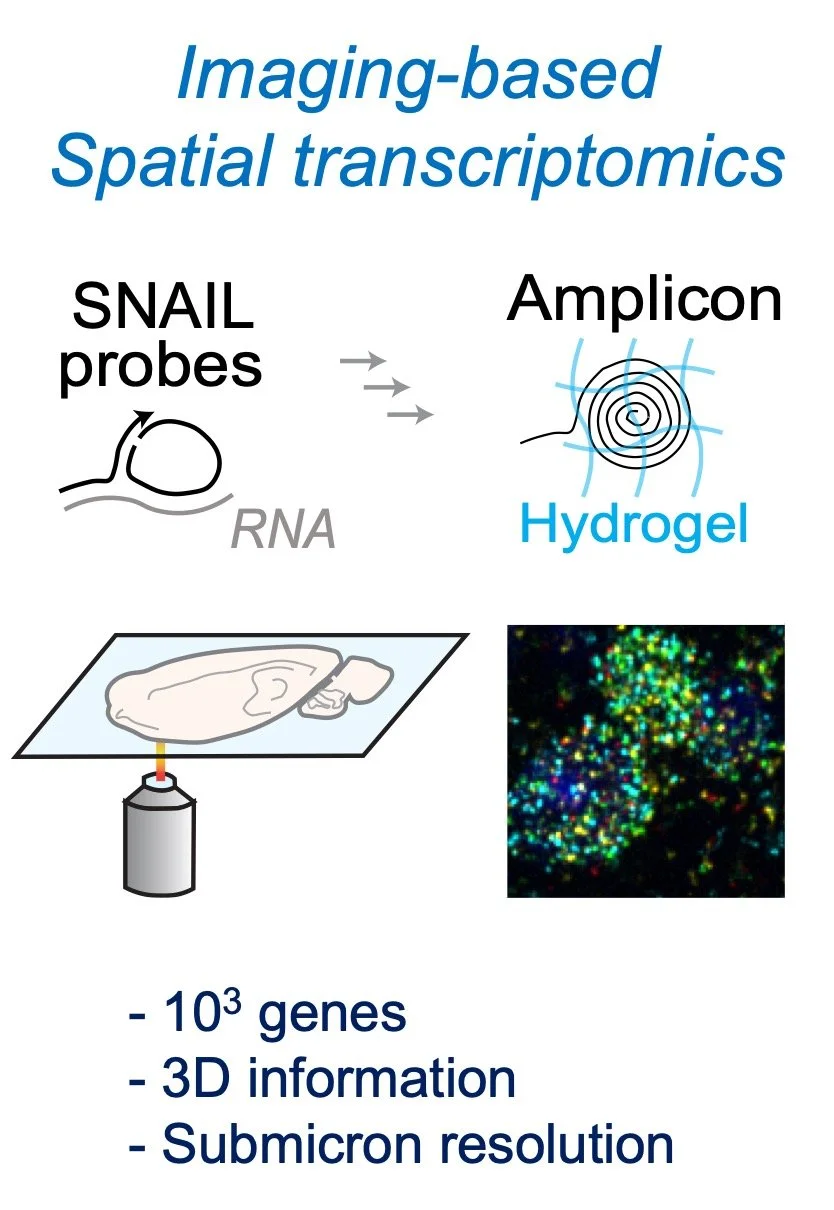
Building on imaging-based spatial transcriptomics tools, we aim to enable tools to characterize various aspects of RNA biology in situ in complex tissues.
Molecular Tissue Biology
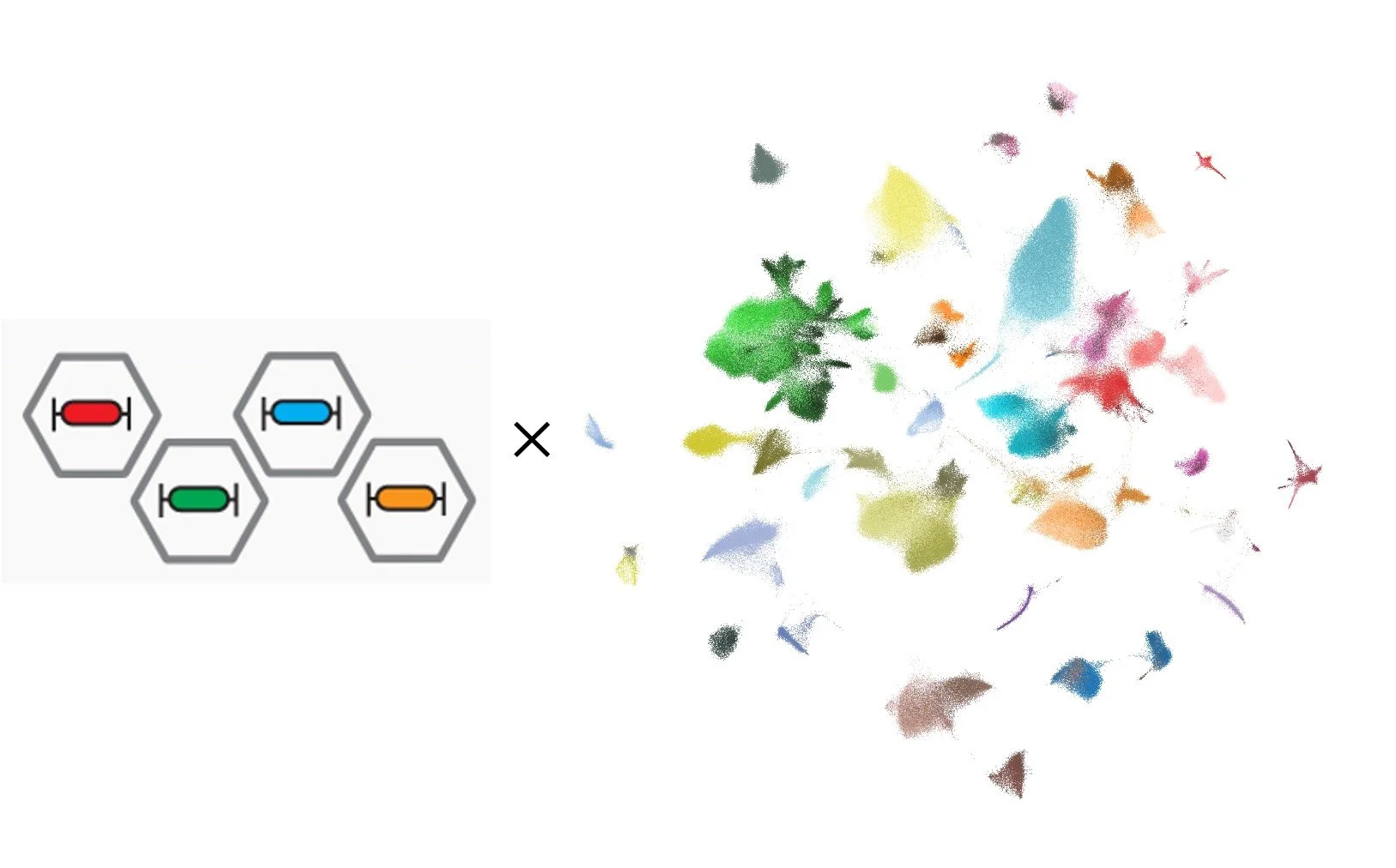
Spatial cell typing of mouse brain slices with 1,022 genes measured at the molecular resolution. Left: spatial cell type maps; Right: cell-type marker gene expression patterns.
Spatial information provides invaluable context for tissue physiology and pathology. Understanding how cellular neighborhoods evolve surrounding disease hallmarks can provide insights into disease diagnosis and prognosis.
Spatial omics technology captures gene expression information while maintaining the native tissue environment, bridging genomics and histology. For example, spatial transcriptomics allows for identifying molecular cell types and molecular tissue regions in addition to tissue anatomy.
We aim to understand the molecular foundations of brain health and disease by combining spatial molecular mapping with phenotypic and functional explorations, such as cell differentiation and migration during brain development and region-specific vulnerability of neurodegenerative diseases.
Delivery and Perturbation
Effective and cell-type-specific gene delivery remains highly desired for both fundamental studies of gene functions and clinical applications. Spatial profiling of gene delivery vectors in situ opens opportunities to characterize vector biodistribution, expression, and cellular impact in specific cell types and tissue microenvironments.
We aim to enable multiplexed in situ evaluation of various carriers (such as viral vectors, nanoparticles, engineered viral-like vesicles, and cell-based therapies), achieving co-profiling of the perturbation/therapeutics distribution and responses for mechanistic disease understandings and therapeutics optimization.